Does mRNA technology based COVID vaccine (Pfizer & Moderna) already contain DNA? Plasmids? Kevin McKernan? Is the finding by Aldén et al. thus non-consequential, alarming as it was that mRNA was
reverse transcribed; results indicate up-take of Pfizer into human liver cell line Huh7, leading to changes in LINE-1 expression & distribution; evidence Pfizer mRNA is reverse transcribed in 6 hours
Yet if there is already DNA contamination of the Pfizer & Moderna mRNA technology vaccines, then what are the implications? What exactly have we put into our bodies? Is out genetic material being re-programmed? Why? Was this really a binary bioweapon with stage one now delivered into us?
See Aldén et al. above:
‘In this study, we investigated the effect of BNT162b2 on the human liver cell line Huh7 in vitro. Huh7 cells were exposed to BNT162b2, and quantitative PCR was performed on RNA extracted from the cells. We detected high levels of BNT162b2 in Huh7 cells and changes in gene expression of long interspersed nuclear element-1 (LINE-1), which is an endogenous reverse transcriptase. Immunohistochemistry using antibody binding to LINE-1 open reading frame-1 RNA-binding protein (ORFp1) on Huh7 cells treated with BNT162b2 indicated increased nucleus distribution of LINE-1. PCR on genomic DNA of Huh7 cells exposed to BNT162b2 amplified the DNA sequence unique to BNT162b2. Our results indicate a fast up-take of BNT162b2 into human liver cell line Huh7, leading to changes in LINE-1 expression and distribution. We also show that BNT162b2 mRNA is reverse transcribed intracellularly into DNA in as fast as 6 h upon BNT162b2 exposure.’
Yet see this substack by ANANDAMIDE (Nepetalactone Newsletter) about Pfizer and Moderna bivalent vaccines containing 20-35% expression vector and are transformation competent in E.coli.
‘
Preamble- Follow on estimations of DNA/RNA contamination with alternative methods and more lots has reproduced contamination above the EMA limit but are less extreme than 20-35%. Please be certain to read the follow on substacks. These substacks look at Qubit readings and qPCR and found 43:1 - 161:1 Ratio of RNA:DNA in the Pfizer monovalent vaccines. Limitations- All lots evaluated to date are expired and this could lead to faster RNA decay than DNA decay. It is also possible, the Agilent Tape station DNA gels are staining some RNA and inflating the DNA estimates in the below study.
Previous work demonstrated that the Pfizer and Moderna bivalent vaccines are contaminated with the expression vectors used to manufacture the mRNAs. While EMA documents suggest these double stranded DNAs (dsDNAs) are linearized, no data is provided to quantitate this important step. Linear plasmids are less replication competent than circular plasmids. With early estimates of billions of potentially contaminating molecules of DNA, even 99% efficient linearization reactions could still leave millions of replication competent plasmids behind.
The initial sequencing of the mRNA vaccines did not sequence deeply enough to ascertain the completeness of the linearization reaction. The initial sequencing survey of these vaccines also utilized a directional RNA-Seq library construction method that contained actinomycin D. This additive is used to suppress DNA amplification in the RNA-Seq method as it intercalates with DNA and suppresses DNA dependent RNA polymerase activity of many polymerases. Thus the original 1:350 -1:3000 DNA:RNA read count levels likely under estimate the level of plasmid contamination present in the Moderna and Pfizer bivalent vaccines.
We explored several methods to properly assess the ratio of circular to linear DNA contaminating the vaccines. One method utilized RNase A treatment of the vaccine to remove the modified mRNA. These RNase A treated samples were then DNA purified. Once we had DNA from the vaccine, we could evaluate it with multiple orthogonal assays to triangulate a quantitative measurement of the nucleic acids.
Method 1) Transform vaccine derived DNA into Kanamycin sensitive E.coli cells and plate on Kanamycin selective plates. Circular plasmid will provide antibiotic resistance to E.coli and thus present colonies on Kan plates. Linear DNA is less likely to provide antibiotic selection.
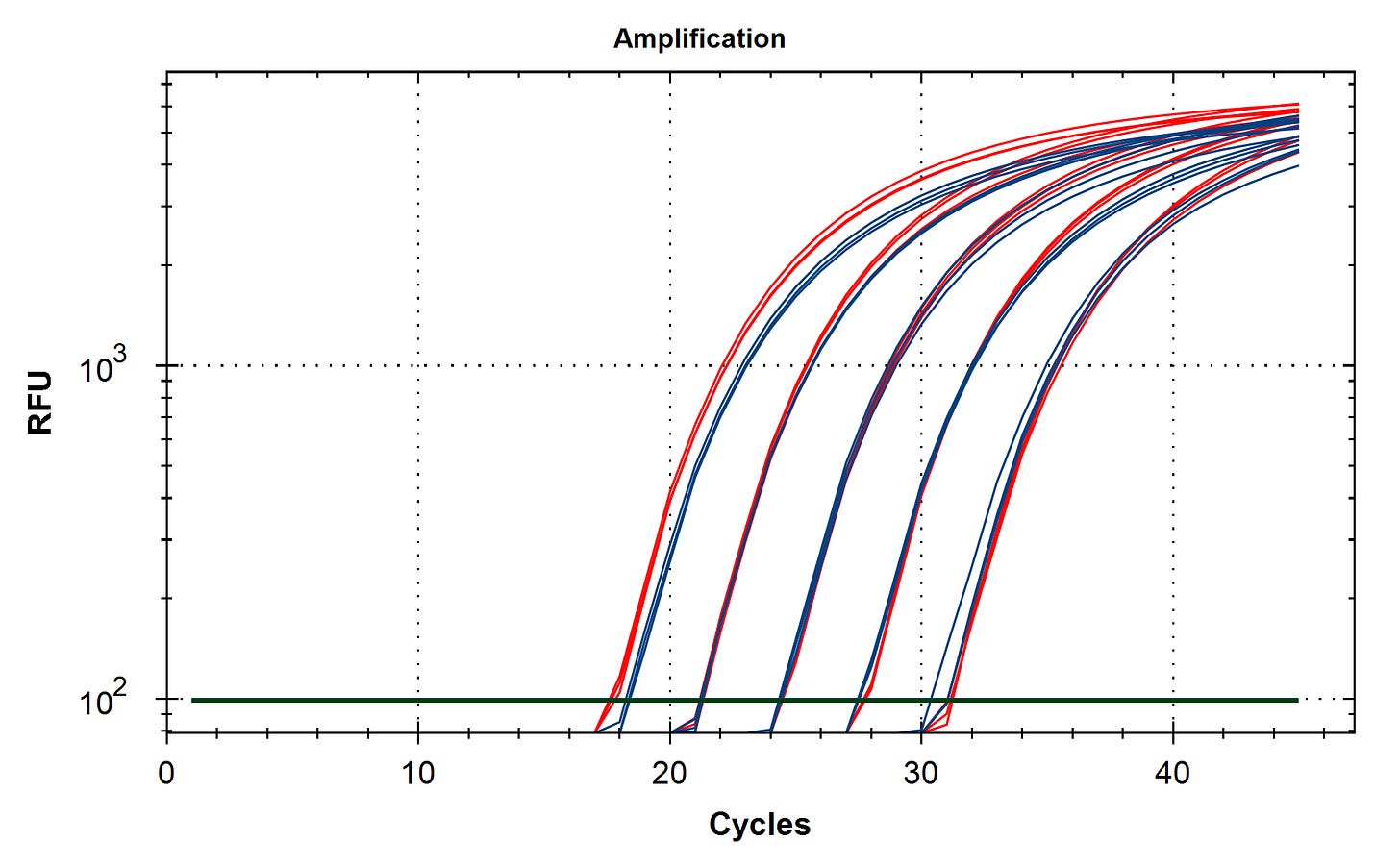
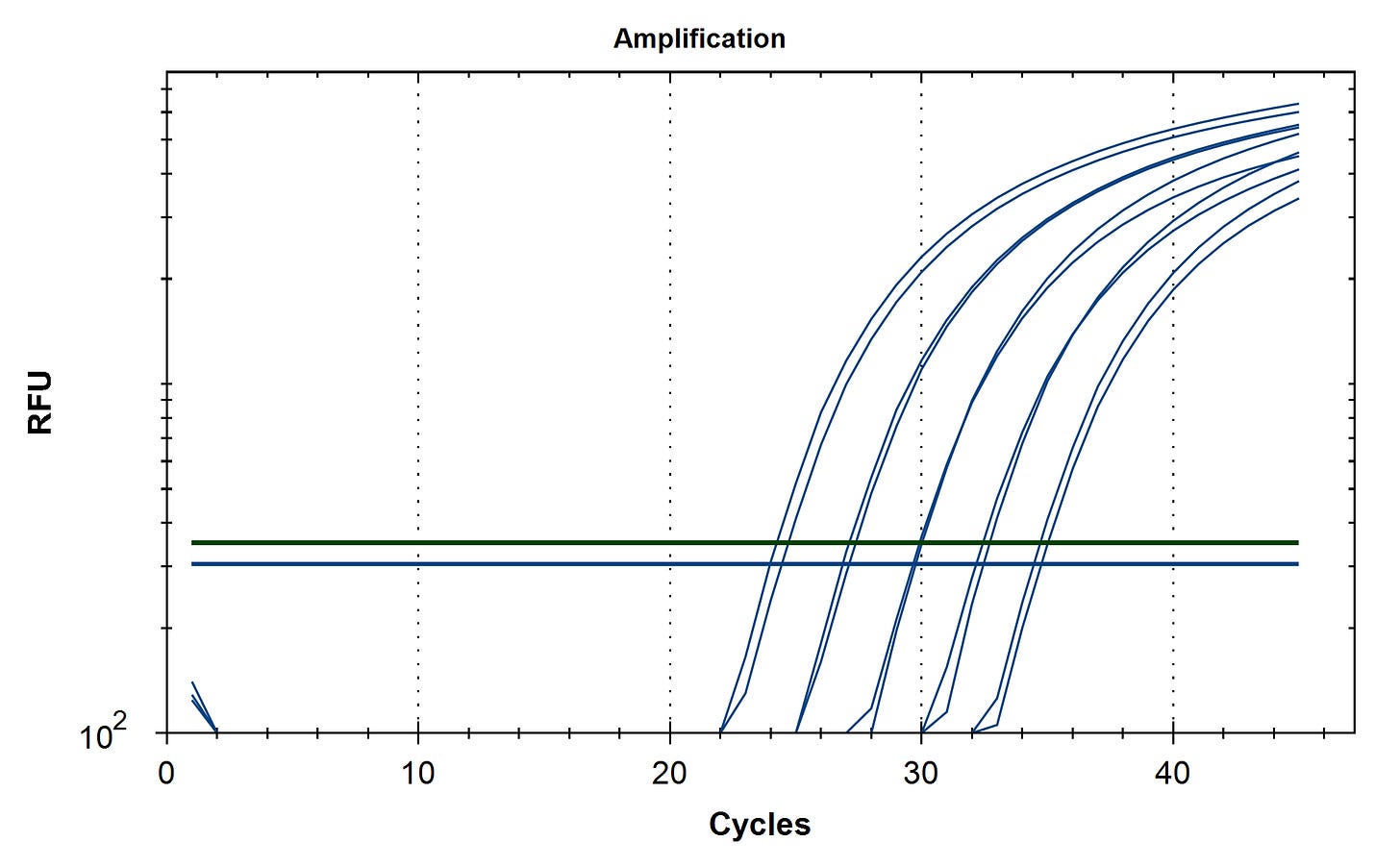
Method 2 ) Design a multiplexed qPCR assay that targets the vector and the spike mRNA insert. These can be used to estimate the plasmid to mRNA ratio more quantitatively than actinomycin D based RNA-sequencing kits that bias the sequencing reads towards RNA.
Method 3) Electrophoresis of purified DNA and RNA from the vaccines to estimate the relative amounts and size of each nucleic acid.
Method 4) Whole genome shotgun sequence the DNA to afford higher coverage over the vector DNA than was obtainable with mRNA present in the library. The 2 million fold sequencing coverage over the vaccine only provided 500-4000 fold coverage over the vector. If we remove the RNA and focus the sequencers on the just the DNA, we can obtain 2 million fold coverage over the plasmid to look for rare circles.
Introduction
The manufacturing of vaccine based mRNAs is best described by Nance et al.
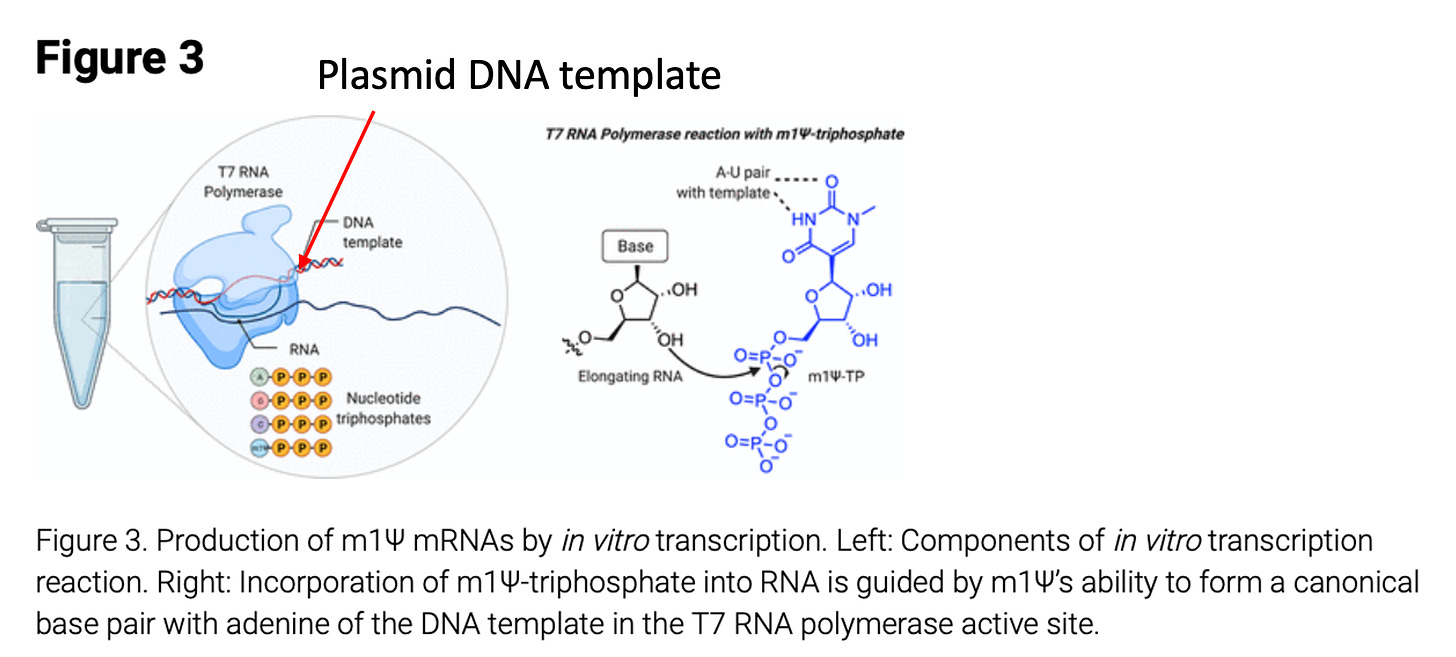
This process can lead to residual plasmid DNA being left in the vaccine mRNAs. The method can also lead to truncated mRNA synthesis as seen in many RNA integrity (RIN plots) data listed as a concern by the EMA and TGA. The presence of E.coli based plasmids is a canary for Lipopolysaccharide (LPS or endotoxin) contamination. Whenever you see high levels of plasmid contamination derived from gram negative bacteria like E.coli, you should expect high levels of endotoxin contamination. Injecting endotoxin can lead to anaphylaxis and toxic shock syndrome.
The vector sequences discovered in previous sequencing work do not contain the long Poly A tracts described by Nance et al. This implies the poly-adenylation is performed after mRNA synthesis. This could be achieved with a polyA polymerase or a RNA ligase. The later method is more compatible with the unique poly-A tail described in Nance et al.
However, RNA ligases would also poly-adenylate truncated mRNAs from the original T7 polymerase manufacturing step. The polyadenlyation of truncated mRNAs would ensure the translation of truncated proteins. These questions should be further explored as poly A tracts are also notorious regions for assembly error. (See March 10th update below).
Methods
RNase A treatment of the Vaccines
RNase A cleaves both uracils and cytosines. N1-methylpseudouridine is known to be RNAse-L resistant but RNase A will cleave cytosines which still exist in the mRNAs. This leaves predominantly DNA for sequencing. Vaccine mRNA that was previously sequenced and discussed here, was treated at 37C for 20 minutes with 3ul of 20 Units/ul Monarch RNase A from NEB. The RNase reaction was purified using 1.5X of SenSATIVAx (Medicinal Genomics #420001). Sample were eluted in 20ul ddH20 after DNA purification. 15ul was used for DNA sequencing and 5ul used for transformation of E.coli.
Transformation of E.coli
50ul of NEBExpress competent E.coli cells (NEB#C2523I) were heat shocked at 42C for 20 seconds with 5ul of DNA (RNase treated vaccine) according to the manufacturers instructions. Cells were recovered in SOC for 1 hour with shaking at 37C. 200-800ul of SOC was plated on Kanamycin 25 plates (Teknova) and grown at 37C for 20 hours. Images were taken at both 20 hours and 44 hours.
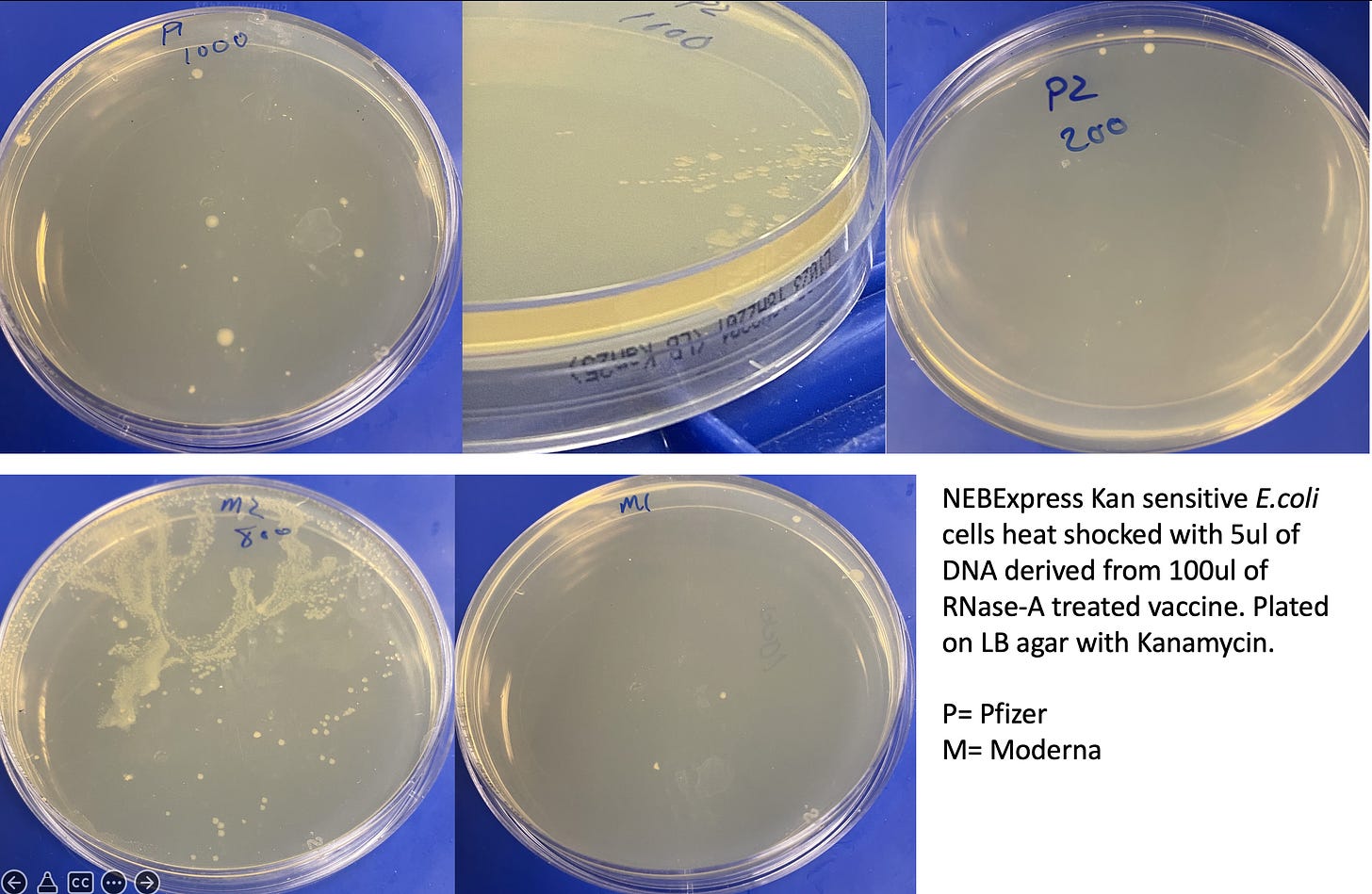
Colonies were picked into 250ul of ddH20 and lysed with 2% LiDS at 37C for 5 minutes. DNA was purified with 1.5X Ampure and 2x 70% EtOH washes. DNA was run on a Agilent Tape station to assess gDNA and presence of plasmids. E.coli qPCR (Medicinal Genomics) was utilized to confirm the colonies were in fact E.coli.
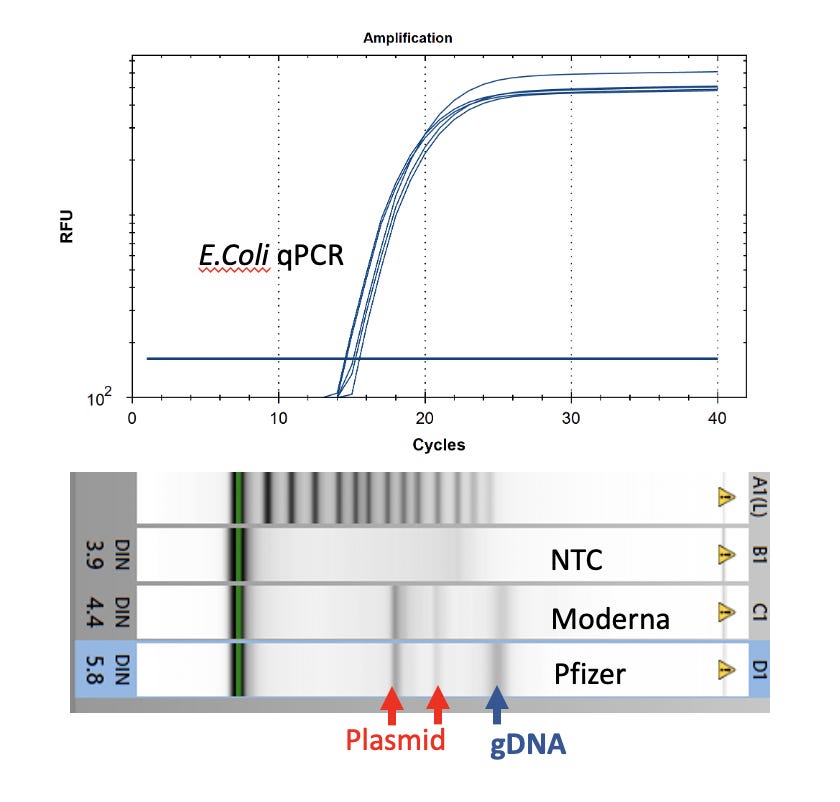
Watchmaker Genomics fragment libraries were constructed from these E.coli colonies for further sequence confirmation of the transformation.
Whole genome shotgun of Pre-transformation RNase’d Vaccines.
15ul of the DNA (prior to E.coli transformation) was converted into sequence ready libraries using Watchmakers Genomics WGS library construction kit.
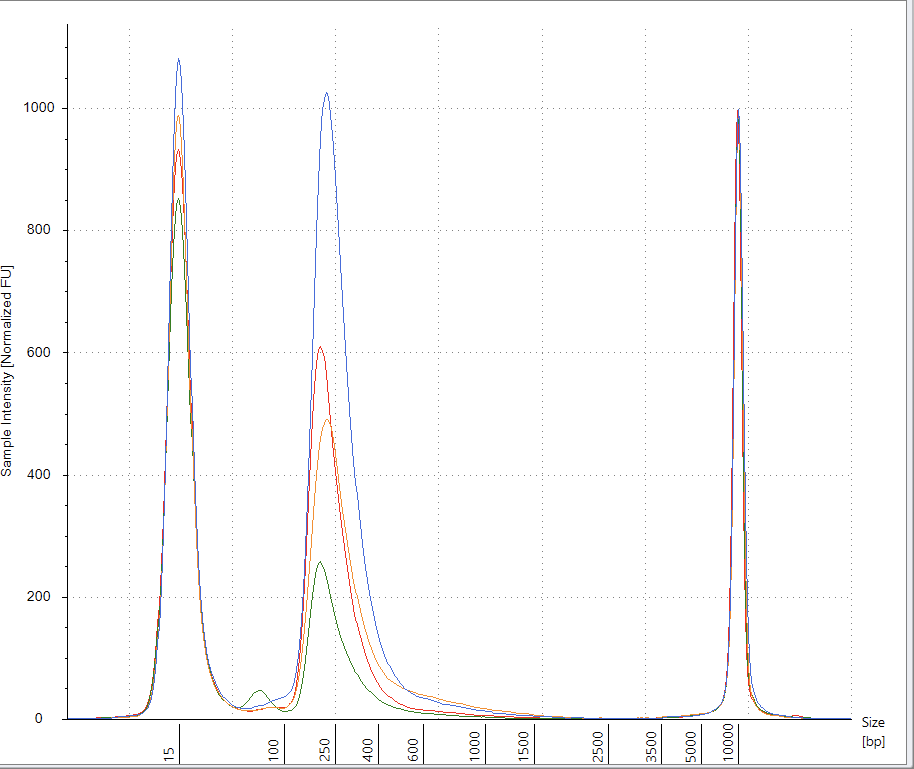
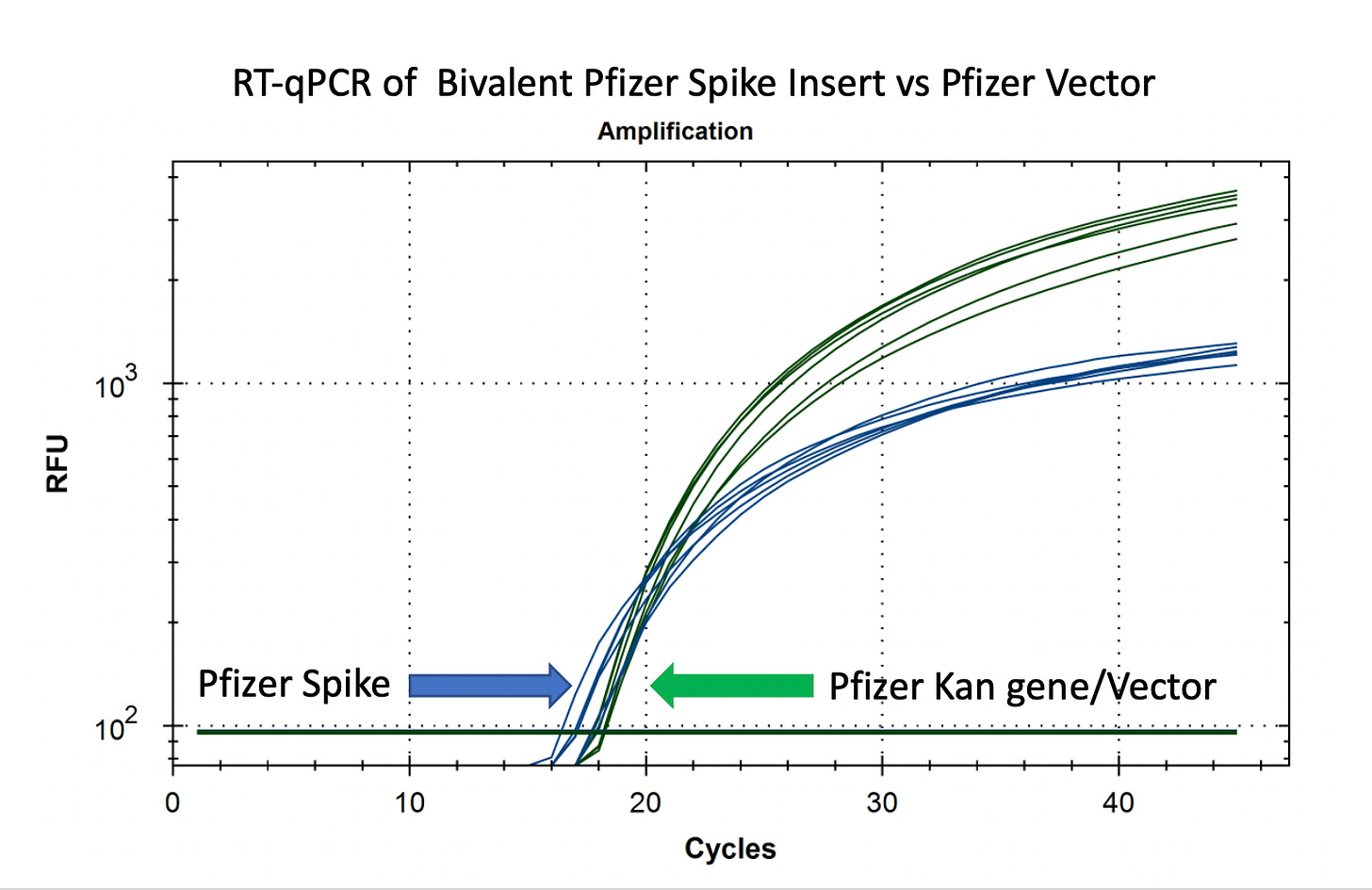
RT-qPCR of plasmid and insert of mRNA vaccines prior to transformation
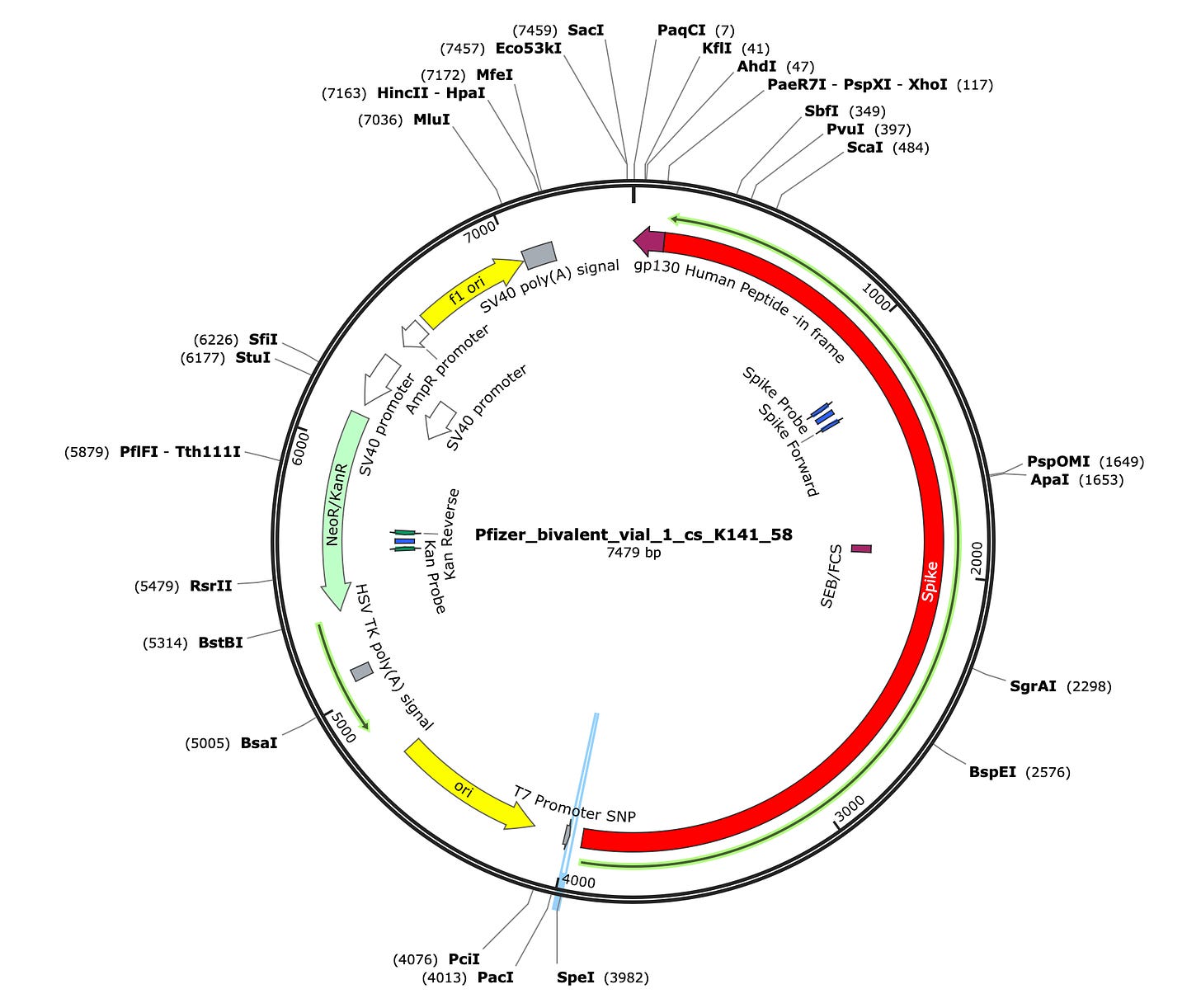
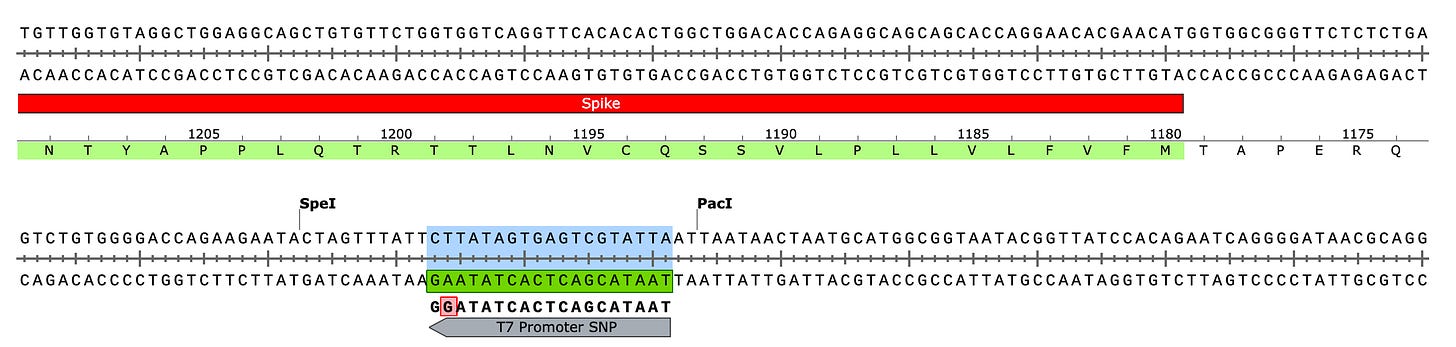
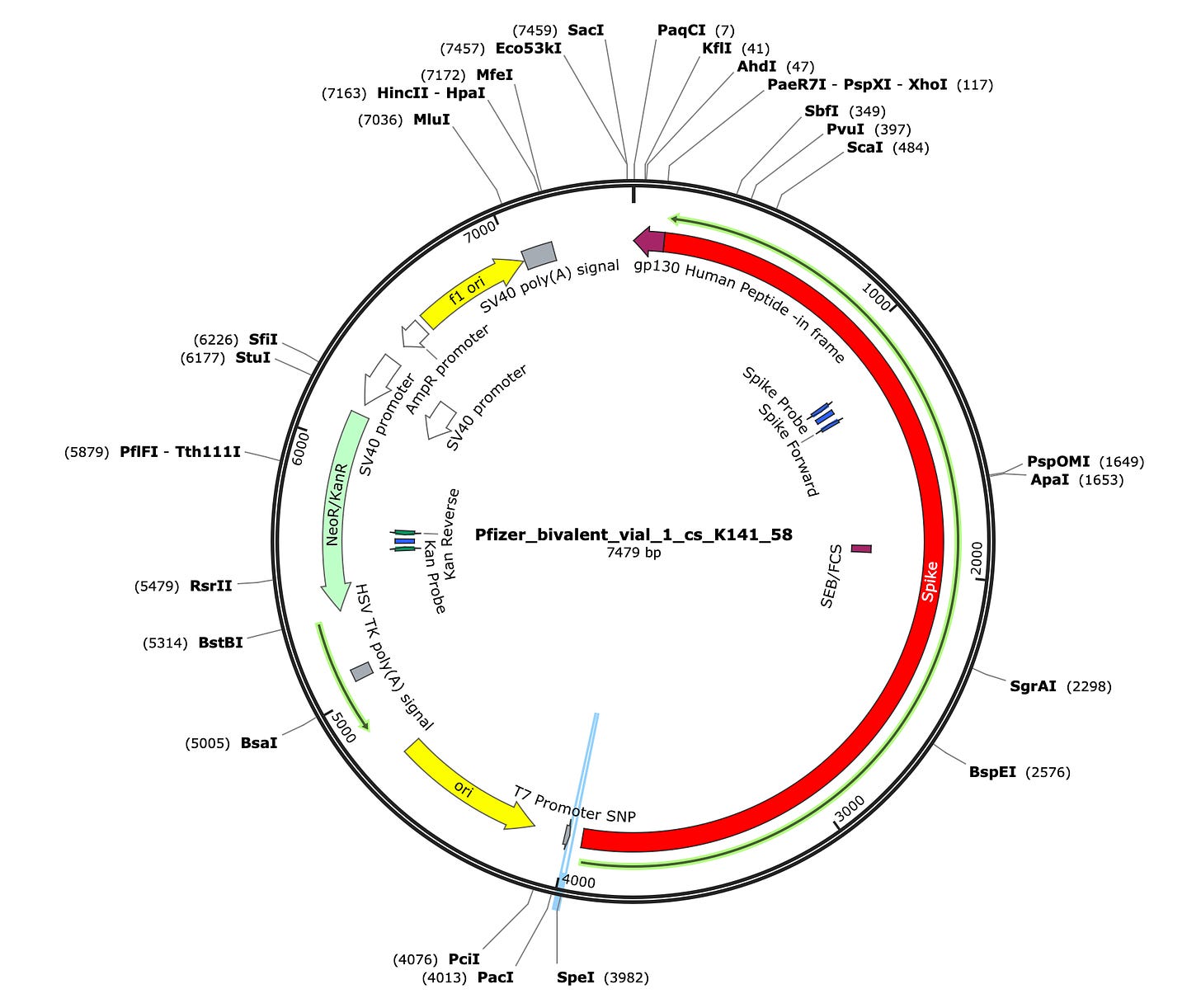
Due to the use of actinomycin D in the RNA-seq first strand synthesis, RT-qPCR and qPCR was used to assess the insert to plasmid ratio. qPCR primers were designed using IDTs Primer Quest software targeting the Kanamycin gene in the plasmid (HEX) and the Spike protein in the plasmid and the mRNA (FAM). The primer coordinates are depicted in vector map produced by SnapGene.
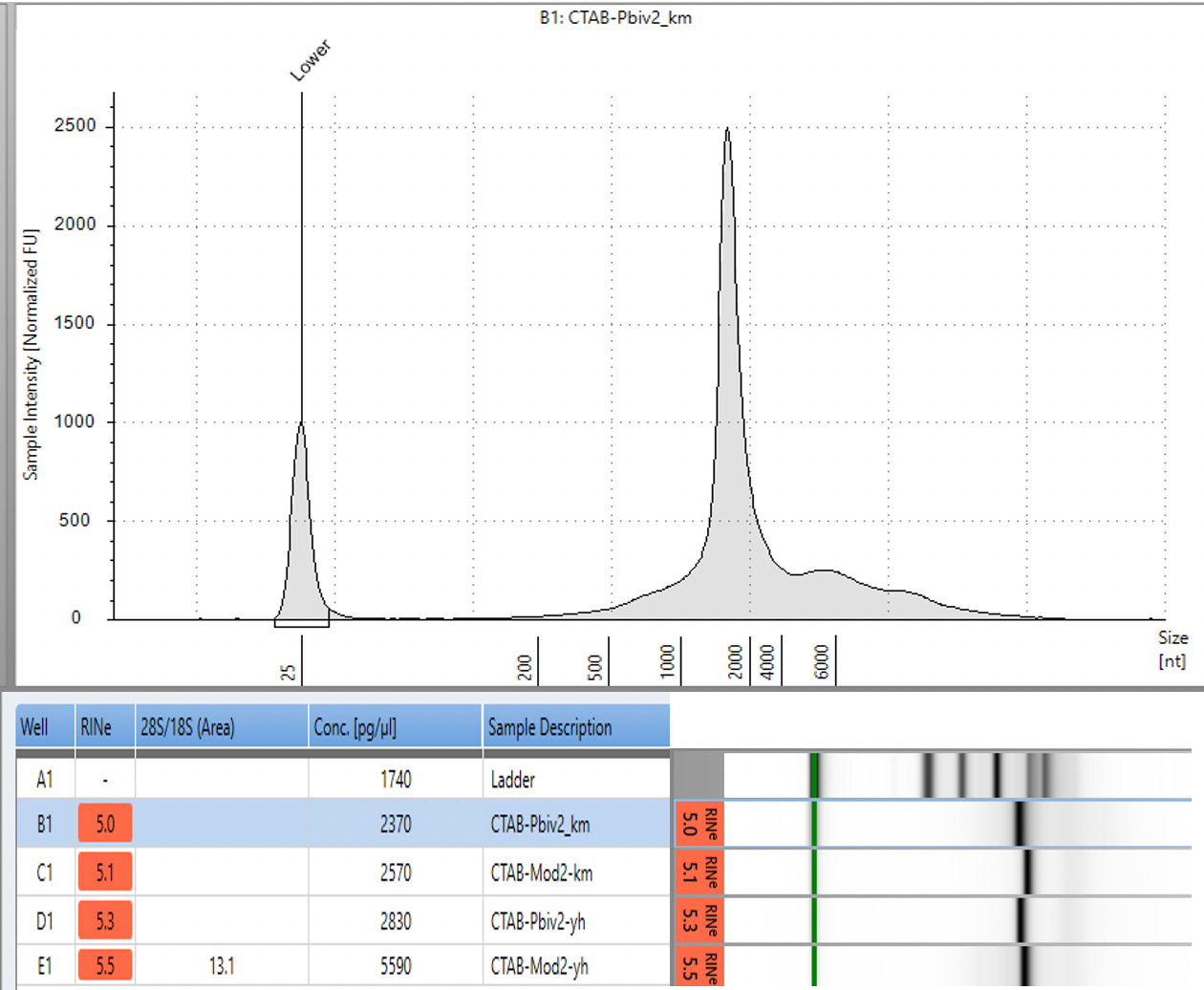
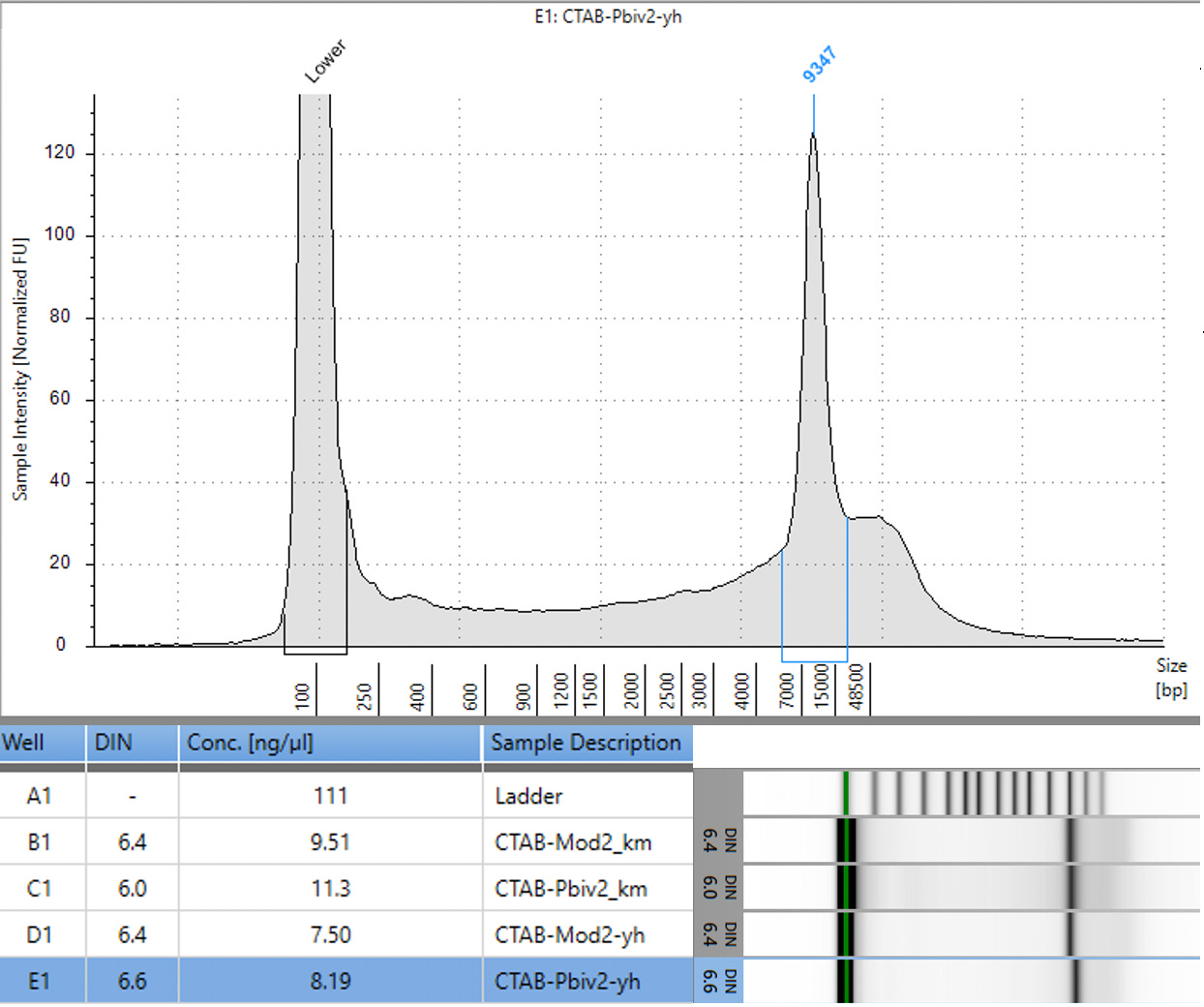
CTAB/Chloroform/SPRI purification of Vaccines
Some variability in qPCR performance was noted with our LiDs/SPRI purification method of the vaccines. This left some samples opaque and is believed to be residual LNPs in the purification. A CTAB/Chloroform/SPRI isolation was optimized to address this and used for further qPCR and Agilent electrophoresis in Figures 10, 11 and 12. Briefly, 300ul of Vaccine was added to 500ul of CTAB (MGC solution A in SenSATIVAx MIP purification kit. #420004). The sample was then vortexed and heated for 5 minutes at 37C. 800ul of chloroform was added, vortexed and spun at 19,000 rpms for 3 minutes. The top 250ul of aqueous phase was collected and added to 250ul of solution B and 1ml of magnetic binding buffer. Samples were vortexed and incubated for 5 minutes and magnetically separated. The supernatant was removed and the beads washed with 70% Ethanol two times. Samples were finally eluted in 300ul of MGC elution buffer.
Conclusions
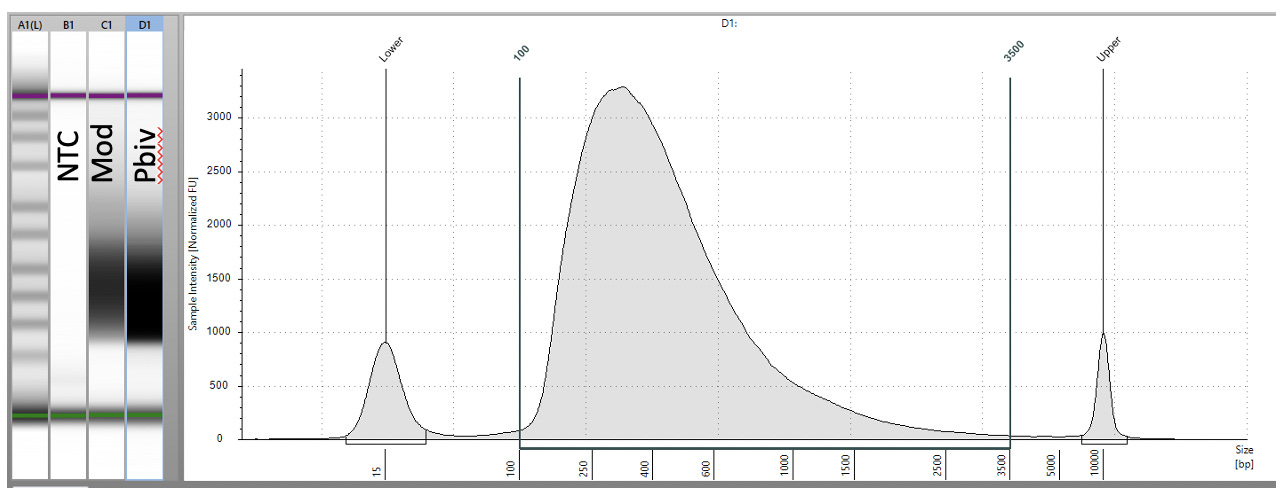
Previous RNA-Seq based estimates of the double stranded DNA contamination in the vaccines significantly under reported the magnitude of the contamination. Using qPCR and electrophoresis, we demonstrate the dsDNA contamination levels are 100 fold higher and imply trillions of DNA molecules per dose. The Pfizer DNA contamination ranges from 8.19-11.3 ng/ul with 23-28ng/ul of mRNA. The Moderna DNA contamination ranges from 7.5 - 9.5ng/ul with 25.7ng/ul - 55.9ng/ul of mRNA. This averages to 9.1ng/ul mean DNA concentration versus 33.4ng/ul mean RNA concentration.
This equates to 27.3% (9.1/33.4) of the nucleic acid in each vaccine being expression vector. This is several orders of magnitude over the the EMAs limit of 330ng/mg.
An unknown portion of these dsDNA contaminants are replication competent plasmids that can transform E.coli with a simple 20 second 42C heat shock treatment. These plasmids provide antibiotic resistance on LB-Kan plates and can be isolated from E.coli cultures. It is unlikely these plasmids will express spike protein in non-laboratory modified E.coli as the ribosomal signals in the vaccine mRNA are designed for mammalian translation. The T7 promoter is known to leak in mammalian cell lines and some laboratory E.coli genotypes but is not expected to leak in wild type E.coli. This may enable mRNA to be expressed from these plasmids in mammalian cells but unless the plasmids are integrated into the human genome, they are unlikely to be replicated to high copy number.
While bacteria are unlikely to express this spike protein, bacteria can replicate this plasmid and serve as a bactofection source for introduction of these mammalian expression plasmids to human cells.
Given the near equimolar contamination, studies evaluating the reverse transcriptase capacity of LINE-1 should be reconsidered. If each injection provides trillions of dsDNA contaminants, LINE-1 RT activity is not a necessary step for genome integration. The critiques of Alden et al focused primarily on the fact that LINE-1 is predominantly expressed in cancer cells lines and that the LINE-1 observation shouldn’t be extrapolated to patients. The vaccines are providing trillions of dsDNAs containing a potentially leaky T7 promoter encoding a spike protein with a Kozak consensus sequence. With these levels of contamination, RT activity from LINE-1 is not a prerequisite for genome integration. These data cannot inform on genome integration and further IRB reviewed deep sequencing work is required to address rare mosaic integration events in patients. Regardless of these hypothetical concerns, the dsDNA contamination exceeds the EMA specifications by several orders of magnitude and further scrutiny should be applied to the endotoxin levels and dsRNA levels in these vaccines.
These data are preliminary and further sequencing confirmation will be made publicly available once complete. Quantitative PCR assays are now available for detection of both Moderna and Pfizer monovalent and bivalent mRNA vaccines and their respective contaminating vector DNAs. This assay also detects the Janssen adenovirus spike sequence. These may be helpful for screening blood, semen, serum and other tissues for blood blanks and fertility clinics concerned with vaccine contamination.
Comments regarding Peer Review- This article will be critiqued for not submitting to the church of academic gatekeeping (Peer Review). The pandemic has revealed many of the warts of this process. You can read about how we can transform peer review with Bitcoin on this substack. While these platforms are being built, the best one can do publishing early and preliminary work is to propose and enable rapid review by the marketplace. Presenting scientific work that contains an easy to verify or falsify hypothesis is the key to migrating to a more decentralized science. This work was careful to present a hypothesis that can be confirmed or falsified in a high school biology lab. All that is needed is an electrophoresis assay that measures RNA and DNA and these results can be readily reproduced. The market will validate this finding long before traditional peer review even puts its boots on. Independent wet lab reproduction trumps 3 anonymous readers every time.
Update March 10th.
While it is notable that the vector has a short poly A tract, we should wait until the ‘RNase A’ sequencing libraries are back before to we speculate too much about the manufacturing process ligating on poly A tracts. The RNase A libraries focus the sequencer on the DNA in the vaccine and allow us to examine those molecules without the 1000X higher signal from the mRNAs.
It is an important area of focus as it may be critical for interpreting the smeary western blots discussed by the EMA and others. Public sequence QC of the plasmids or the vaccine lots would greatly reduce public anxiety over this topic. We once enjoyed rapid data release policies in genomics with the Human genome project and the Bermuda accord. Some of this spirit persists today with over 6.7 millions SARs-CoV-2 genomes public in NCBI. Vaccine genomics, however defies this scientific sprit.
The vaccine sequences to date can be counted on 1 hand and can only be found in the depths of github or substack. A few accidental vaccine genomes exist due to sequencing patients plasma but there is no concerted effort to shine a spot lot on the variability of this novel genomic manufacturing process.
We have sequences of the viral genomes that involuntarily enter our body but none on the ones we can actually control via injection? The fidelity of the ExoN/RdRp polymerase gene in SARs-CoV-2 is higher than the error prone T7 polymerase with N1 methylpseudouridine suggesting sequence surveillance of the vaccine supply chain is more important than the sequencing the virus. This is a incoherent position given the RO of the vaccine traversed the globe via social media transmission as quickly as a biological virus.
Why caution is in order-
Poly A tracts are notorious locations for error in assembly as they have low complexity and often little signature to algorithmically organize. Sequences are assembled from pairs of 150 base pair reads. Imagine a jigsaw puzzle where all the pieces are the same color. You really only have the edge pieces to work with and in the case of sequencing a mixture of RNA and DNA you have conflicting edge pieces (at the poly A tail) that want to break the puzzle into two different puzzles.
The plasmid DNA sequences being circular, will disagree the most with the mRNA sequences right at the end of the poly A. So this may be an assembly artifact that DNA only sequencing clarifies. More to come on the next stack.
EMA documentation on 330ng/mg DNA/RNA limit. Page 74.’
Follow the work of Kevin McKernan@Kevin_McKernan
anandamide.substack.com




Yes, in vitro reverse transcription was known more than a year ago. In vivo reverse transcription should have been published by now, but human beings - especially doctors, with all their training - are so stupid. Ah well, play stupid games, win stupid prizes. Get your clots, nerve palsies, heart attacks, strokes, sudden deaths, infertility, cancers, etc. At this stage, it's karma.
This is very good and very alarming research and it needs to be published in a scientific journal ASAP. Let the peer reviewers come at it. The problem is finding a journal that actually is "scientific." Do those even exist any more,? Many appear to present ideology masquerading as science and straight out propaganda. If this was to be published in a journal it would probably be retracted within days.